
This AOP is licensed under the BY-SA license. This license allows reusers to distribute, remix, adapt, and build upon the material in any medium or format, so long as attribution is given to the creator. The license allows for commercial use. If you remix, adapt, or build upon the material, you must license the modified material under identical terms.
AOP: 360
Title
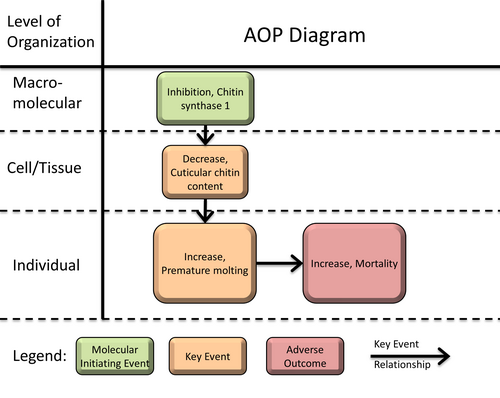
Chitin synthase 1 inhibition leading to mortality
Short name
Graphical Representation
Point of Contact
Contributors
- Simon Schmid
- You Song
- Knut Erik Tollefsen
Coaches
- Shihori Tanabe
OECD Information Table
| OECD Project # | OECD Status | Reviewer's Reports | Journal-format Article | OECD iLibrary Published Version |
|---|---|---|---|---|
| 1.94 | WPHA/WNT Endorsed | Scientific Review | iLibrary link |
This AOP was last modified on April 29, 2023 16:03
Revision dates for related pages
| Page | Revision Date/Time |
|---|---|
| Inhibition, Chitin synthase 1 | February 24, 2021 04:41 |
| Decrease, Cuticular chitin content | February 17, 2021 05:37 |
| Increase, Premature molting | February 17, 2021 05:30 |
| Increase, Mortality | October 26, 2020 05:18 |
| Inhibition, CHS-1 leads to Decrease, Cuticular chitin content | February 17, 2021 07:50 |
| Decrease, Cuticular chitin content leads to Increase, Premature molting | February 17, 2021 08:20 |
| Increase, Premature molting leads to Increase, Mortality | February 17, 2021 08:47 |
| Polyoxin B | May 24, 2018 15:54 |
| Polyoxin D | October 23, 2020 06:20 |
| Nikkomycins | May 24, 2018 15:54 |
| Captan | October 23, 2020 06:50 |
| Captafol | October 23, 2020 06:52 |
| Folpet | October 23, 2020 06:53 |
Abstract
In order to grow and develop, arthropods need to shed their exoskeleton (or cuticle) periodically and replace it with a new one in a process called molting. Successful molting, and therefore a successful development necessitates stability and integrity of the cuticle to support muscular contractions involved in the shedding of the old cuticle. The integrity of the cuticle is largely dependent on the N-acetylglucosamine (GlcNAc) polymer chitin. Therefore, arthropods heavily rely on chitin synthesis as chitin is one of the main constituents of the cuticle. The cuticular chitin synthase (CHS-1) is the key enzyme in the biosynthetic pathway and arthropods are therefore especially dependent on its proper function. The present AOP describes the effects of chemical inhibition of the cuticular chitin synthase (CHS-1) on the molting process leading to increased mortality in arthropods. Inhibition of CHS-1 is the molecular initiating event and leads to a decreased chitin content in the arthropod cuticle which leaves the organism immature at the stage for ecdysis. This phenomenon can be described as premature molting. The organism eventually dies due to being stuck in the old cuticle or due to the consequences of a weak exoskeleton after ecdysis. The AOP is considered to be very consistent. Essentiality of key events was rated as high for every key event and the biological plausibility was rated as high for the whole AOP. However, there does not exist very much empirical evidence that allows to draw a representative conclusion on dose concordance along the AOP whereas time concordance can be supported by knockdown studies of CHS-1. Therefore, empirical evidence was considered to be moderate and the quantitative understanding was considered to be low. The overall confidence in the AOP was valued as moderate. The present AOP will guide assay development for further experimental studies by revealing data and knowledge gaps. One of its primary applications will also be providing guidance in screening strategies in order to identify chemicals directly interacting with CHS-1.
AOP Development Strategy
Context
Arthropods (including insects, crustaceans and arachnids) need to shed their exoskeleton in order to grow and reproduce. This process, also called molting or ecdysis, is mediated by behavioural mechanisms which involve the skeletal muscles (Ayali 2009; Song et al. 2017a). In order to properly shed its cuticle, the organism needs to possess a newly synthesized cuticle that possesses a certain integrity to support this process. Since chitin is a major constituent of the cuticle, it contributes substantially to its integrity (Cohen 2001; Vincent and Wegst 2004). Chitin is synthesized from uridine diphosphate-N-Acetylglucosamine (UDP-GlcNAc) in a polymerization reaction by the transmembrane enzyme chitin synthase isoform 1 (CHS-1). CHS-1 is localized on the apical side in the cuticular epithelium. Since chitin and the process of chitin synthesis does not occur in vertebrates, it can and has been exploited for the design of pest controlling agents. Inhibitors of chitin synthesis may not only be of use for the control of unwanted arthropods and fungi, they may also pose a risk for beneficial arthropods such as insects and crustaceans. Disruption of chitin synthesis or the endocrine mechanisms controlling molting generally lead to a disruption of ecdysis (Merzendorfer et al. 2012; Song et al. 2017a; Song et al. 2017b). If the amount of chitin in the cuticle decreases, the affected organism may not be able to molt properly and will most probably die of starvation or suffocation (Camp et al. 2014; Song et al. 2017a). Alternatively, if molting is completed despite an immature cuticle, the organism may be deformed and die as a consequence of a weak cuticle. Therefore, the present AOP should build the basis of a mechanistic approach for the systematic evaluation and the risk assessment of chemicals interfering with chitin synthesis by directly inhibiting CHS-1.
Strategy
Summary of the AOP
Events:
Molecular Initiating Events (MIE)
Key Events (KE)
Adverse Outcomes (AO)
| Type | Event ID | Title | Short name |
|---|
| MIE | 1522 | Inhibition, Chitin synthase 1 | Inhibition, CHS-1 |
| KE | 1523 | Decrease, Cuticular chitin content | Decrease, Cuticular chitin content |
| KE | 1524 | Increase, Premature molting | Increase, Premature molting |
| AO | 350 | Increase, Mortality | Increase, Mortality |
Relationships Between Two Key Events (Including MIEs and AOs)
| Title | Adjacency | Evidence | Quantitative Understanding |
|---|
| Inhibition, CHS-1 leads to Decrease, Cuticular chitin content | adjacent | Moderate | Low |
| Decrease, Cuticular chitin content leads to Increase, Premature molting | adjacent | Moderate | Low |
| Increase, Premature molting leads to Increase, Mortality | adjacent | Moderate | Low |
Network View
Prototypical Stressors
Life Stage Applicability
| Life stage | Evidence |
|---|---|
| Larvae | High |
| Juvenile | High |
| Adult | Moderate |
Taxonomic Applicability
| Term | Scientific Term | Evidence | Link |
|---|---|---|---|
| Pieris brassicae | Pieris brassicae | High | NCBI |
| Anopheles gambiae | Anopheles gambiae | High | NCBI |
| Lucilia cuprina | Lucilia cuprina | High | NCBI |
| Tribolium castaneum | Tribolium castaneum | High | NCBI |
| Bombyx mori | Bombyx mori | High | NCBI |
| Anopheles quadrimaculatus | Anopheles quadrimaculatus | High | NCBI |
| Trichoplusia ni | Trichoplusia ni | High | NCBI |
| Artemia salina | Artemia salina | High | NCBI |
| Daphnia magna | Daphnia magna | High | NCBI |
| Hyalophora cecropia | Hyalophora cecropia | High | NCBI |
| Ostrinia nubilalis | Ostrinia nubilalis | High | NCBI |
| Bradysia hygida | Bradysia hygida | Moderate | NCBI |
| Mamestra brassicae | Mamestra brassicae | Moderate | NCBI |
| Chilo suppressalis | Chilo suppressalis | Moderate | NCBI |
| Locusta migratoria | Locusta migratoria | Moderate | NCBI |
| Nilaparvata lugens | Nilaparvata lugens | Moderate | NCBI |
| Aphis glycines | Aphis glycines | Moderate | NCBI |
| Lepeophtheirus salmonis | Lepeophtheirus salmonis | Moderate | NCBI |
| Panonychus citri | Panonychus citri | Moderate | NCBI |
| Grapholita molesta | Grapholita molesta | Moderate | NCBI |
| Ectropis obliqua | Ectropis obliqua | Moderate | NCBI |
| Tigriopus japonicus | Tigriopus japonicus | Moderate | NCBI |
Sex Applicability
| Sex | Evidence |
|---|---|
| Unspecific | Moderate |
Overall Assessment of the AOP
Domain of Applicability
Taxonomic: Since the whole phylum of arthropods is dependent on the synthesis of chitin to molt successfully, it is extremely likely that the AOP is applicable to all arthropods. Effect data along the AOP exist from Dipteran, Lepidopteran and Coleopteran insect species as well as from Branchiopods and Anostracans of the crustacea. Although data is limited, KEs seem to be well conserved across taxa, as shown in available studies with specific stressors known to inhibit CHS and in studies where CHS-1 was knocked down by RNA interference. However, due to limited data availability, it was not possible to cover whole taxa but rather single species in the assessment of KEs. Alignment of amino acid residues in the catalytic center of CHS-1 using the Sequence Alignment to Predict Across Species Susceptibility tool (SeqAPASS, https://seqapass.epa.gov/seqapass, LaLone et al. 2016), confirmed structural and functional conservation in various insect, arachnid and crustacean species, strenghtening the evidence for the applicability domain to be the whole phylum of arthropods. However, taxonomic applicability may not only be defined by structural conservation of the protein sequence. So the evidence for the taxonomic applicability for species with support only from sequence alignment was judged as moderate, whereas evidence for species with support from sequence alignment and effect data was judged as high.
Life stage: The AOP is applicable for organisms undergoing continuous molt cycles. As insects do not molt in their adulthood, the AOP is only applicable for larval and pupal stages of insects. Crustaceans and arachnids grow and molt throughout their lifetime (Passano 1961; Uhl et al. 2015), which makes the AOP applicable to all life stages, where juvenile life stages might be more susceptible to chemical perturbations due to higher growth rate and therefore more frequent molting.
Sex: The AOP is applicable to all sexes.
Chemical: Substances known to trigger the MIE and leading to the AO are of the family of pyrimidine nucleosides (e.g. polyoxin D, polyoxin B and nikkomycin Z) (Osada 2019). There also exists evidence for phthalimides (captan, captafol and folpet) to inhibit CHS-1 activity and to decrease the cuticular chitin content in vitro (Cohen and Casida 1982; Gelman and Borkovec 1986). However, as these substances are known to covalently bind to thiol groups in proteins (Lukens and Sisler 1958), it is not clear if the inhibition is due to specific CHS-1 inhibition or due to unspecific protein binding.
Essentiality of the Key Events
The essentiality of all key events was considered as high. Essentiality evaluations were mainly based on specifically designed studies demonstrating the expected effect pattern predicted by the AOP to occur after knockdown of CHS-1.
Inhibition, Chitin synthase 1 (High): Knockdown of the cuticular chitin synthase leads to the expected pattern of effects described in this AOP. It decreases the cuticular chitin content and leads to premature molting associated mortality in insects (Arakane et al. 2005; X. Zhang et al. 2010; Li et al. 2017; Zhai et al. 2017). If the cuticular chitin content was not directly measured as endpoint, knockdown of the CHS-1 led directly to the occurrence of premature molting associated increase of mortality (Chen et al. 2008; X. Zhang et al. 2010; Wang et al. 2012; Yang et al. 2013; Shang et al. 2016; Mohammed et al. 2017; Wang et al. 2019; Ye et al. 2019; Ullah et al. 2020)
Decrease, Cuticular chitin content (High): Abolishment of the cuticular chitin synthesis through knockdown of CHS-1 leads to premature molting associated mortality (Arakane et al. 2005; X. Zhang et al. 2010; Li et al. 2017; Zhai et al. 2017). By knocking down the UDP-GlcNAc pyrophosphorylase (UAP), which catalyzes the last sugar conversion before the polymerization to chitin, it was shown that reduced chitin content leads to the same outcome as the knockdown of CHS-1. Namely premature molting and increased mortality (Arakane et al. 2011; Liu et al. 2013). Knockdown of trehalase genes, which constitutes the start of the chitin synthetic pathway and convert trehalose to glucose, leads to a similar pattern of effects, namely decreased cuticular chitin content and premature molting associated mortality (Chen et al. 2010; Shi et al. 2016).
Increase, Premature molting (High): Several studies show that premature molting is a direct consequence of decreased chitin synthesis and leads to increased mortality. The KE is consistently listed as cause for mortality when CHS-1 is knocked down throughout a number of studies (Arakane et al. 2005; Chen et al. 2008; J. Zhang et al. 2010; X. Zhang et al. 2010; Wang et al. 2012; Yang et al. 2013; Shang et al. 2016; Li et al. 2017; Mohammed et al. 2017; Zhai et al. 2017; Wang et al. 2019; Ye et al. 2019).
Increase, Mortality (High): Increased mortality was observed in all of the abovementioned studies.
Evidence Assessment
Biological Plausibility: The biosynthesis of chitin is well characterized and is conserved among arthropods. Although the exact mode of action of chitin synthases remains elusive, it is widely accepted and well established that the chitin synthase is the key enzyme in the pathway, polymerizing chitin using UDP-N-Acetylglucosamine as substrate (Merzendorfer and Zimoch 2003). Arthropod cuticles mostly consist of chitin embedded into a matrix of cuticular proteins. It is therefore widely accepted that chitin contributes crucially to the quality and function of the cuticle (Reynolds 1987; Muthukrishnan et al. 2012). The molting process requires the new cuticle to be strong enough to withstand the stresses of ecdysis. During ecdysis, arthropods pause food intake and growth. If ecdysis is initiated before the new cuticle is strong enough, the organism likely dies of starvation or growth arrest (Song, Villeneuve, et al. 2017). It was also reported that certain arthropods pause respiration during ecdysis, which may lead to suffocation (Camp et al. 2014). Based on the well-established biological knowledge on the processes this AOP bases on, the biological plausibility for all KER was rated as high.
Empirical Evidence: Empirical evidence assessment was conducted on the basis of in vitro and in vivo experiments performed with stressors affecting key events throughout the AOP. Studies showed that the key events are affected by model stressors such as Polyoxin D and Nikkomycin Z, which are able to competitively inhibit CHS1 (Endo et al. 1970). Several studies provide evidence that polyoxin B, polyoxin D and nikkomycin Z trigger the MIE in cell free systems of coleopteran, lepidopteran and dipteran insect species (Cohen 1982; Turnbull and Howells 1982; Kuwano and Cohen 1984; Cohen and Casida 1990; Zhang and Yan Zhu 2013). Also the cuticular chitin content was shown to be decreased by polyoxin D and nikkomycin Z in lepidopteran and dipteran species as well as in the crustacean Artemia salina (Gijswijt et al. 1979; Calcott and Fatig 1984; Gelman and Borkovec 1986; Zhuo et al. 2014). The AO is supported by in vivo studies with polyoxin D and nikkomycin Z in dipteran insects and Daphnia magna (Tellam et al. 2000; Tellam and Eisemann 2000; Zhu et al. 2007; Zhang and Yan Zhu 2013; New Zealand Environmental Protection Authority 2015). A major data gap constitutes the absence of data covering the KE “Increase, premature molting”. This KE is mentioned in some studies but never assessed as an individual endpoint (Gijswijt et al. 1979; Tellam et al. 2000). Another major data gap is the lacking quantitative data for KERs. As endpoints were only measured as individual endpoints and not in sequence, it makes it nearly impossible to evaluate the dose for the KEs and KERs. However, data from studies where CHS-1 was knocked down are able to support temporal concordance for all KERs. Knockdown of CHS-1 led to decreased chitin content and subsequently to premature molting associated mortality (Arakane et al., 2005; Li et al., 2017). Based on the major data gaps and therefore the lacking information on dose concordance as well as the given time concordance, empirical evidence was evaluated to be moderate for the whole AOP.
Overall confidence in the AOP: Both, essentiality of KEs and the biological plausibility of the whole AOP were considered to be high. However, due to missing quantitative data and the lack of evidence for dose concordance, empirical evidence was judged to be moderate. Therefore the overall confidence in the AOP was evaluated as moderate.
Known Modulating Factors
Quantitative Understanding
Quantitative data are limited for all KER and therefore the whole AOP. Therefore, predictions on the occurrence of downstream KE and the AO on the basis of the occurrence of upstream KEs is not readily feasible. Quantitative understanding of the AOP was therefore considered to be low.
Considerations for Potential Applications of the AOP (optional)
Arthropods are responsible for many functions in terrestrial as well as aquatic ecosystems and are therefore jointly responsible for ecosystem health (Seastedt and Crossley 1984; Losey and Vaughan 2006; LeBlanc 2007). Therefore, it is important to develop AOPs which enhance the mechanistic knowledge on chemicals, such as chitin synthesis inhibitors, which may pose a risk to non-target arthropods. Those AOPs will contribute to the systematic use of mechanistic data to preserve beneficial arthropod populations and ecosystem health. The present AOP will help to guide future experimental studies by identifying data gaps. This will lead to the identification and development suitable bioassays in order to populate the AOP with (quantitative) experimental data which may allow for predictions of regulatory relevant endpoints on the basis of the occurrence of the MIE. The present AOP may also guide screening strategies in order to identify chemicals inhibiting CHS-1. The identified substances may then be prioritized and undergo a thorough hazard assessment. As there already exist approaches to assess mixture toxicity using the AOP framework (Altenburger et al. 2012; Beyer et al. 2014), the present AOP could be employed for the effect assessment of mixtures of chemicals that share the same KEs (e.g. AOP #361, aopwiki.org/aops/361, AOP #358, aopwiki.org/aops/358, and AOP #359, aopwiki.org/aops/359).
References
Altenburger R, Scholz S, Schmitt-Jansen M, Busch W, Escher BI. 2012. Mixture toxicity revisited from a toxicogenomic perspective. Environ Sci Technol. 46(5):2508–2522. doi:10.1021/es2038036.
Arakane Y, Baguinon MC, Jasrapuria S, Chaudhari S, Doyungan A, Kramer KJ, Muthukrishnan S, Beeman RW. 2011. Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochem Mol Biol. 41(1):42–50. doi:10.1016/j.ibmb.2010.09.011. http://dx.doi.org/10.1016/j.ibmb.2010.09.011.
Arakane Y, Muthukrishnan S, Kramer KJ, Specht CA, Tomoyasu Y, Lorenzen MD, Kanost M, Beeman RW. 2005. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol Biol. 14(5):453–463. doi:10.1111/j.1365-2583.2005.00576.x.
Ayali A. 2009. The role of the arthropod stomatogastric nervous system in moulting behaviour and ecdysis. J Exp Biol. 212(4):453–459. doi:10.1242/jeb.023879.
Beyer J, Petersen K, Song Y, Ruus A, Grung M, Bakke T, Tollefsen KE. 2014. Environmental risk assessment of combined effects in aquatic ecotoxicology: A discussion paper. Mar Environ Res. 96:81–91. doi:10.1016/j.marenvres.2013.10.008. http://dx.doi.org/10.1016/j.marenvres.2013.10.008.
Calcott PH, Fatig RO. 1984. Inhibition of Chitin metabolism by Avermectin in susceptible Organisms. J Antibiot (Tokyo). 37(3):253–259. doi:10.7164/antibiotics.37.253.
Camp AA, Funk DH, Buchwalter DB. 2014. A stressful shortness of breath: Molting disrupts breathing in the mayfly Cloeon dipterum. Freshw Sci. 33(3):695–699. doi:10.1086/677899.
Chen Jie, Tang B, Chen H, Yao Q, Huang X, Chen Jing, Zhang D, Zhang W. 2010. Different functions of the insect soluble and membrane-bound trehalase genes in chitin biosynthesis revealed by RNA interference. PLoS One. 5(4). doi:10.1371/journal.pone.0010133.
Chen X, Tian H, Zou L, Tang B, Hu J, Zhang W. 2008. Disruption of Spodoptera exigua larval development by silencing chitin synthase gene A with RNA interference. Bull Entomol Res. 98(6):613–619. doi:10.1017/S0007485308005932.
Cohen E. 1982. In vitro chitin synthesis in an insect: formation and structure of microfibrils. Eur J Cell Biol. 26(2):289–294.
Cohen E. 2001. Chitin synthesis and inhibition: A revisit. Pest Manag Sci. 57(10):946–950. doi:10.1002/ps.363.
Cohen E, Casida JE. 1982. Properties and inhibition of insect integumental chitin synthetase. Pestic Biochem Physiol. 17(3):301–306. doi:10.1016/0048-3575(82)90141-9.
Cohen E, Casida JE. 1990. Insect and Fungal Chitin Synthetase Activity: Specificity of Lectins as Enhancers and Nucleoside Peptides as Inhibitors. Pestic Biochem Physiol. 37(3):249–253. doi:10.1016/0048-3575(90)90131-K.
Endo A, Kakiki K, Misato T. 1970. Mechanism of action of the antifugal agent polyoxin D. J Bacteriol. 104(1):189–196. doi:10.1128/jb.104.1.189-196.1970.
Gelman DB, Borkovec AB. 1986. The pharate adult clasper as a tool for measuring chitin synthesis and for identifying new chitin synthesis inhibitors. Comp Biochem Physiol Part C, Comp. 85(1):193–197. doi:10.1016/0742-8413(86)90073-3.
Gijswijt MJ, Deul DH, de Jong BJ. 1979. Inhibition of chitin synthesis by benzoyl-phenylurea insecticides, III. Similarity in action in Pieris brassicae (L.) with Polyoxin D. Pestic Biochem Physiol. 12(1):87–94. doi:10.1016/0048-3575(79)90098-1.
Kuwano E, Cohen E. 1984. The use of a Tribolium chitin synthetase assay in studying the effects of benzimidazoles with a terpene moiety and related compounds. Agric Biol Chem. 48(6):1617–1620. doi:10.1080/00021369.1984.10866362.
LaLone, C.A., Villeneuve, D.L., Lyons, D., Helgen, H.W., Robinson, S.L., Swintek, J.A., Saari, T.W., Ankley, G.T., 2016. Sequence alignment to predict across species susceptibility (seqapass): A web-based tool for addressing the challenges of cross-species extrapolation of chemical toxicity. Toxicol. Sci. 153, 228–245. https://doi.org/10.1093/toxsci/kfw119
LeBlanc GA. 2007. Crustacean endocrine toxicology: A review. Ecotoxicology. 16(1):61–81. doi:10.1007/s10646-006-0115-z.
Li T, Chen J, Fan X, Chen W, Zhang W. 2017. MicroRNA and dsRNA targeting chitin synthase A reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens. Pest Manag Sci. 73(7):1529–1537. doi:10.1002/ps.4492.
Liu X, Li F, Li D, Ma E, Zhang W, Zhu KY, Zhang J. 2013. Molecular and functional analysis of UDP-N-acetylglucosamine Pyrophosphorylases from the Migratory Locust, Locusta migratoria. PLoS One. 8(8). doi:10.1371/journal.pone.0071970.
Losey JE, Vaughan M. 2006. The economic value of ecological services provided by insects. Bioscience. 56(4):311–323. doi:10.1641/0006-3568(2006)56[311:TEVOES]2.0.CO;2.
Lukens RJ, Sisler HD. 1958. 2-Thiazolidinethione-4-carboxylic acid from the reaction of captan with cysteine. Science (80- ). 127(3299):650. doi:10.1126/science.127.3299.650.
Merzendorfer H, Kim HS, Chaudhari SS, Kumari M, Specht CA, Butcher S, Brown SJ, Robert Manak J, Beeman RW, Kramer KJ, et al. 2012. Genomic and proteomic studies on the effects of the insect growth regulator diflubenzuron in the model beetle species Tribolium castaneum. Insect Biochem Mol Biol. 42(4):264–276. doi:10.1016/j.ibmb.2011.12.008. http://dx.doi.org/10.1016/j.ibmb.2011.12.008.
Merzendorfer H, Zimoch L. 2003. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 206(24):4393 LP – 4412. doi:10.1242/jeb.00709. http://jeb.biologists.org/content/206/24/4393.abstract.
Mohammed AMA, DIab MR, Abdelsattar M, Khalil SMS. 2017. Characterization and RNAi-mediated knockdown of Chitin Synthase A in the potato tuber moth, Phthorimaea operculella. Sci Rep. 7(1):1–12. doi:10.1038/s41598-017-09858-y. http://dx.doi.org/10.1038/s41598-017-09858-y.
Muthukrishnan S, Merzendorfer H, Arakane Y, Kramer KJ. 2012. Chitin Metabolism in Insects. Elsevier B.V. http://dx.doi.org/10.1016/B978-0-12-384747-8.10007-8.
New Zealand Environmental Protection Authority. 2015. Application for approval to import ESTEEM for release. https://www.epa.govt.nz/assets/FileAPI/hsno-ar/APP202334/fbce9a39e6/APP202334-APP202334-Staff-Report-Final-updated.pdf.
Osada H. 2019. Discovery and applications of nucleoside antibiotics beyond polyoxin. J Antibiot (Tokyo). 72(12):855–864. doi:10.1038/s41429-019-0237-1. http://dx.doi.org/10.1038/s41429-019-0237-1.
Passano LM. 1961. The regulation of crustacean metamorphosis. Integr Comp Biol. 1(1):89–95. doi:10.1093/icb/1.1.89.
Reynolds SE. 1987. The cuticle, growth and moulting in insects: The essential background to the action of acylurea insecticides. Pestic Sci. 20(2):131–146. doi:10.1002/ps.2780200207.
Seastedt TR, Crossley DA. 1984. Influence of on arthropods ecosystems. Bioscience. 34(3):157–161.
Shang F, Xiong Y, Xia WK, Wei DD, Wei D, Wang JJ. 2016. Identification, characterization and functional analysis of a chitin synthase gene in the brown citrus aphid, Toxoptera citricida (Hemiptera, Aphididae). Insect Mol Biol. 25(4):422–430. doi:10.1111/imb.12228.
Shi JF, Xu QY, Sun QK, Meng QW, Mu LL, Guo WC, Li GQ. 2016. Physiological roles of trehalose in Leptinotarsa larvae revealed by RNA interference of trehalose-6-phosphate synthase and trehalase genes. Insect Biochem Mol Biol. 77:52–68. doi:10.1016/j.ibmb.2016.07.012.
Song Y, Evenseth LM, Iguchi T, Tollefsen KE. 2017b. Release of chitobiase as an indicator of potential molting disruption in juvenile Daphnia magna exposed to the ecdysone receptor agonist 20-hydroxyecdysone. J Toxicol Environ Heal - Part A Curr Issues. 80(16–18):954–962. doi:10.1080/15287394.2017.1352215. https://doi.org/10.1080/15287394.2017.1352215.
Song Y, Villeneuve DL, Toyota K, Iguchi T, Tollefsen KE. 2017a. Ecdysone Receptor Agonism Leading to Lethal Molting Disruption in Arthropods: Review and Adverse Outcome Pathway Development. Environ Sci Technol. 51(8):4142–4157. doi:10.1021/acs.est.7b00480.
Tellam RL, Eisemann C. 2000. Chitin is only a minor component of the peritrophic matrix from larvae of Lucilia cuprina. Insect Biochem Mol Biol. 30(12):1189–1201. doi:10.1016/S0965-1748(00)00097-7.
Tellam RL, Vuocolo T, Johnson SE, Jarmey J, Pearson RD. 2000. Insect chitin synthase. cDNA sequence, gene organization and expression. Eur J Biochem. 267(19):6025–6043. doi:10.1046/j.1432-1327.2000.01679.x.
Turnbull IF, Howells AJ. 1982. Effects of several larvicidal compounds on chitin biosynthesis by isolated larval integuments of the sheep blowfly Lucilia cuprina. Aust J Biol Sci. 35(5):491–504. doi:10.1071/BI9820491.
Uhl G, Zimmer SM, Renner D, Schneider JM. 2015. Exploiting a moment of weakness: Male spiders escape sexual cannibalism by copulating with moulting females. Sci Rep. 5(July):1–7. doi:10.1038/srep16928.
Vincent JFV, Wegst UGK. 2004. Design and mechanical properties of insect cuticle. Arthropod Struct Dev. 33(3):187–199. doi:10.1016/j.asd.2004.05.006.
Wang Y, Fan HW, Huang HJ, Xue J, Wu WJ, Bao YY, Xu HJ, Zhu ZR, Cheng JA, Zhang CX. 2012. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae). Insect Biochem Mol Biol. 42(9):637–646. doi:10.1016/j.ibmb.2012.04.009. http://dx.doi.org/10.1016/j.ibmb.2012.04.009.
Wang Z, Yang H, Zhou C, Yang WJ, Jin DC, Long GY. 2019. Molecular cloning, expression, and functional analysis of the chitin synthase 1 gene and its two alternative splicing variants in the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). Sci Rep. 9(1):1–14. doi:10.1038/s41598-018-37488-5. http://dx.doi.org/10.1038/s41598-018-37488-5.
Yang WJ, Xu KK, Cong L, Wang JJ. 2013. Identification, mRNA expression, and functional analysis of chitin synthase 1 gene and its two alternative splicing variants in oriental fruit fly, Bactrocera dorsalis. Int J Biol Sci. 9(4):331–342. doi:10.7150/ijbs.6022.
Ye C, Jiang Y Di, An X, Yang L, Shang F, Niu J, Wang JJ. 2019. Effects of RNAi-based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum). Sci Rep. 9(1):1–10. doi:10.1038/s41598-019-39837-4. http://dx.doi.org/10.1038/s41598-019-39837-4.
Zhai Y, Fan X, Yin Z, Yue X, Men X, Zheng L, Zhang W. 2017. Identification and Functional Analysis of Chitin Synthase A in Oriental Armyworm, Mythimna separata. Proteomics. 17(21):1–11. doi:10.1002/pmic.201700165.
Zhang J, Liu X, Zhang Jianqin, Li D, Sun Y, Guo Y, Ma E, Zhu KY. 2010. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen). Insect Biochem Mol Biol. 40(11):824–833. doi:10.1016/j.ibmb.2010.08.001. http://dx.doi.org/10.1016/j.ibmb.2010.08.001.
Zhang X, Yan Zhu K. 2013. Biochemical characterization of chitin synthase activity and inhibition in the African malaria mosquito, Anopheles gambiae. Insect Sci. 20(2):158–166. doi:10.1111/j.1744-7917.2012.01568.x.
Zhang X, Zhang J, Zhu KY. 2010. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol. 19(5):683–693. doi:10.1111/j.1365-2583.2010.01029.x.
Zhu KY, Heise S, Zhang J, Anderson TD, Starkey SR. 2007. Comparative Studies on Effects of Three Chitin Synthesis Inhibitors on Common Malaria Mosquito (Diptera: Culicidae). J Med Entomol. 44(6):1047–1053. doi:10.1093/jmedent/44.6.1047.
Zhuo W, Fang Y, Kong L, Li X, Sima Y, Xu S. 2014. Chitin synthase A: A novel epidermal development regulation gene in the larvae of Bombyx mori. Mol Biol Rep. 41(7):4177–4186. doi:10.1007/s11033-014-3288-1.